14 December 2020
With the introduction of opt-in permissions for apps, iOS 14 will make it harder for brands to target consumers on an individual level and to measure results of marketing activities. Bobby Gray, Head of Analytics and Data Marketing at Artefact, considers the impact and explains how brands can respond using first-party data.
Introduction
This project is part of Artefact’s contribution in Tech for Good. The project has been conducted in collaboration with Institut Carnot CALYM, a consortium dedicated to partnership research on lymphoma, and Microsoft.
In autumn 2019, the Institut Carnot CALYM launched a structuring programme aimed at setting up a roadmap to optimise the valorisation and exploitation of data from the clinical, translational and preclinical research conducted by the members of the consortium for more than 20 years. This project, proposed by Pr Camille Laurent (LYSA, IUCT, CHU Toulouse, France) and Pr Christiane Copie (LYSARC, Pierre-Bénite, France), both members of Institut Carnot CALYM, is part of this structuring programme.
The primary objective of this research project is to develop a deep-learning algorithm to assist pathologists in diagnosing Follicular Lymphoma. A secondary objective is to identify informative criteria that could help medical experts understanding the morphological differences between Follicular Lymphoma and Follicular Hyperplasia which will be referred below as FL and FH.
What is Follicular Lymphoma? What are the challenges in its diagnosis?
FL is a subtype of Lymphoma, the most frequent blood cancer in the world. There are more than 80 types of Lymphoma and this diversity makes its diagnosis difficult, even for experts. Moreover, FL is very similar to FH which is not cancerous, adding challenges to its diagnosis.
In this article, we will describe our approach in building a classifier for FL and FH using only labelled whole-slide images. Whole slide images are high resolution digital files of scanned microscope slides. In our case they contain extract of lymph nodes.
How could deep learning help in its detection?
Using whole-slide images of FL and FH, we trained a binary classifier through a patch-based approach. Our model architecture is a simple Resnet-18 trained on a few epochs (~10).
After predicting the class of an observation with the classifier, we extract the last activation layer to build a heatmap on top of the input image to highlight parts that have prompted the model in defining a given class.
Why did we use a patch-based classification?
Patch-based classification is a classification technique where the class of a given observation is built based on the aggregation of the predictions of its components (patches). In our case it is used because the images are way too large to be used directly on the model.
In fact, whole-slide images are very large (~10⁵ pixel square). Their size makes training a deep learning model almost impossible with common tools. To solve this issue, we divided them into patches of the same size following two important criteria:
In patch-based classification, the model output can be interpreted as that of a classical classification except that the first layer of computation is at the whole-slide level. For example, when predicting the class of a slide of FL, a score of 98% would mean that 98 % of the patches it is composed of have been predicted to be FL.
At the dataset level, this slide will be predicted with a score of 0.98 for the FL class.
PS: We made the hypothesis of dividing the images into patches based on medical experts’ conclusions stating that in a whole-slide of FL, the follicles are expected to be present everywhere.
Training Set
Our training set is composed of 58k randomly selected patches (1024 pixel square) of FL and FH extracted from a set of 30 whole-slide images in each of the 2 classes.
Validation Set
20% of the patches was sampled for validating the model performance at training time.
Testing Set
Our testing set is composed of 15 whole-slide images, each divided into patches. This reference set has been used to compare the results of different training approaches that we will precise below.
Modelling
Our testing set is composed of 15 whole-slide images, each divided into patches. This reference set has been used to compare the results of different training approaches that we will precise below.
Before training the deep learning classifier: Image preparation and processing
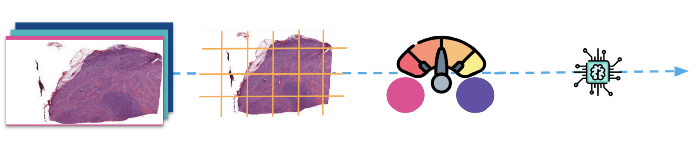
(Above: The images are first divided into patches, then normalised before they are fed to the model for training.)
After training: Inference and interpretation
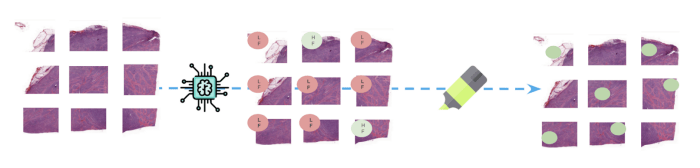
(Above: At inference time, new whole-slide are divided into patches before the model predicts a class for
each one of them. Parts of images responsible for predicting FL class are highlighted to help monitoring
the results.)
In the sections below, we will give the details about these different steps of the pipeline.
Data preparation and processing
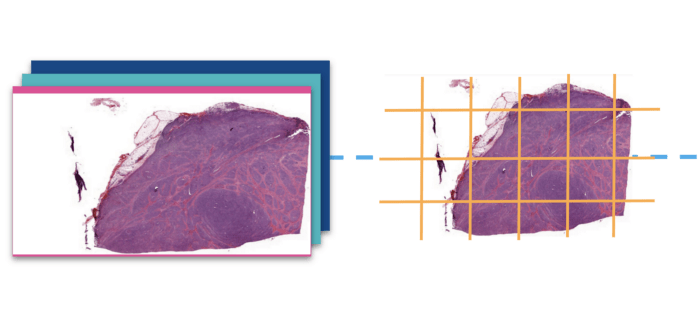
1 — Tiling
As stated earlier, whole-slide images are very large and cannot directly be fed to a classification model unless you are using a super galactic hardware. We used the library openslide to read the slides and its deepzoom support to divide the images into relatively small tiles of size 1024 pixel square. After breaking them into tiles we ran them into a basic cleaner that dropped all tiles that were not at the center of the tissue (borders, holes etc).
2 — Stain normalisation
The second step of our data processing, which is also the most important step, is the stain color normalisation. Staining is the process of highlighting important features on slides and enhancing the contrast between them. The staining system used is the common H&E (Hematoxylin and Eosin).
However, since the images are coming from many different laboratories, we have observed variations in the colouring of the slides. They mainly come from differences in the dying process from one laboratory to another. These differences can affect the model’s performance a lot.
We used classical techniques to normalise the coloration of the dataset before training the model.
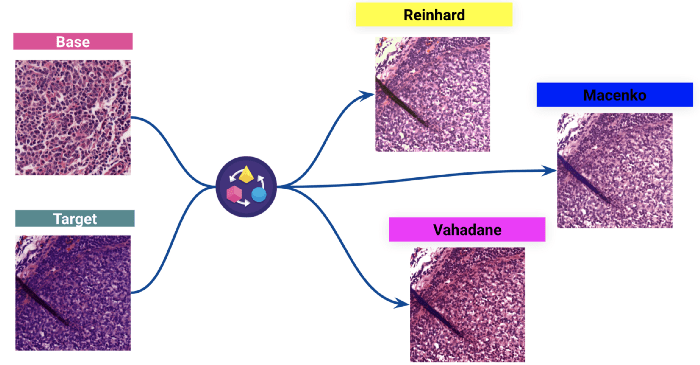
(Above: Results of three different stain normalisation : a target image colouring is normalised to a base image colour distribution.)
We picked the Reinhard technique to see the impact on the model.
Training a Resnet-18 classifier
After processing the whole-slide images, the training went smoothly (dropout, weight decay, etc..). Nothing fancy except from adding mixup in the data augmentation. We used a Resnet18 trained from scratch since pre-trained models were not significantly improving our results. We also preferred the Resnet-18 since the Resnet-34 and Resnet-56 were not improving our performances. After ~10 epochs, our model was ready for testing.
We used the very practical Fastai library to build our models with few efforts.
Testing
The results of 3 experimentation are worth being mentioned:
The results on the test set for these 3 experimentations are shown below:
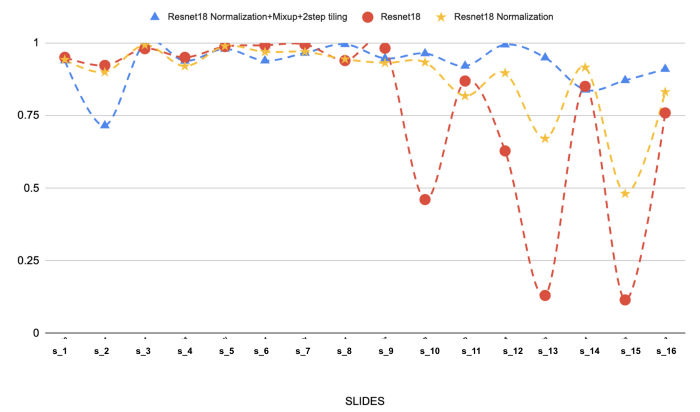
(Above: The results of 3 different models on the 16 selected slides of Follicular Lymphoma. We can see the effect of stain normalisation and mixup on performance.)
Stain normalisation is by far the most important step in our modelling approach. We were experiencing generalisation problems (red line) but it definitely help in solving the issue. Adding mixup and a 2-step tiling makes it even better.
MixUp is a data augmentation technique which consists of creating new observations by linearly interpolating many samples.
Interpreting the results of a computer vision classifier
In order to easily communicate the results to medical experts, we provided images with heatmaps to highlight where the model’s focus was when predicting a given label. We did that by extracting the last activation layer of the convolutional network and by linearly extrapolating it on the image we were predicting onto.
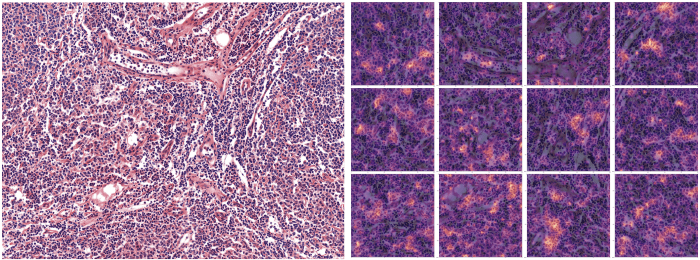
(Above: Parts of the image that has most contributed to the prediction of the class Follicular Lymphoma are highlighted on the right sided image — 12 patches)).
Interpreting the model’s output with heatmaps has been very useful in adjusting the modelling approach as it gives experts ways to analyse what the model is actually doing. Through our exchanges with experts, we (data scientists) were able to adjust how we to handle better the dataset and make the model more robust (i.e able to adapt to different types of inputs). And also to make sure it serves its purpose. It was in fact how we realised the need to normalise the staining of the images.
Conclusion and Key learnings
The goal of this study was to explore the process of creating a good deep learning base classifier for differentiating Follicular Lymphoma and Follicular Hyperplasia. Our keys learnings are listed below:

 BLOG
BLOG